Identification of SIAT7 in Symbiotic Clownfish and a Closely Related Non-Symbiotic Fish
By Deborah Dele-Oni

Portrait of Dele-Oni
Clownfish live in a close symbiotic relationship with sea anemones. This relationship is often used as a teaching tool for students to learn about ecology, evolutionary mutualism, and species interactions. This mutualistic relationship may be due to a sugar the anemones detect in the mucus of the prey species. An enzyme class known as sialyltransferases has been studied because of its importance of sea anemone recognition of prey. This class of sialyltransferases adds chains of sugars to proteins found in mucus. Clownfish may lack a specific type of sialytransferases known as SIAT7, which could be a factor as to why the clownfish do not get stung. However, although SIAT7 was not initially seen does not mean it is not there; rather it could indicate inactivation. Alternatively, clownfish may have SIAT7 in their genomes but may not express it in the cells that make the external mucus. To test this, I am studying both symbiotic and closely related non-symbiotic species to determine if SIAT7 is present in these species. I hypothesize that SIAT7 will be present in both the symbiotic clownfish and non-symbiotic closely related species but is inactive in the skin of symbiotic species. My goals were to test primers on tissues of anemonefish and closely related non-symbiotic species to see where expression occurred.
To accomplish these goals, I will:
- Use degenerate PCR to obtain partial sequences of SIAT7 from symbiotic and non-symbiotic fish species.
- Use inverse PCR to determine the sequences of the regions surrounding the SIAT7 gene.
- Use quantitative PCR to determine which tissues express SIAT7 in symbiotic and non-symbiotic fish species.
To approach this, I knew that SIAT7 had been identified in close relatives of clownfish. If primers were designed based from those sequences and added to DNA of symbiotic clownfish, there would be a product formed if the primers found complementary parts of the DNA. In the spring, I completed degenerate PCR to try and acquire partial sequences of SIAT7 from non-symbiotic fish species. The degenerative primers were created from the bicolor damselfish (Stegastes partitus; Genbank accession XP_008298796.1), and PCR was done on cDNA samples from the ocellaris clownfish (Amphiprion ocellaris) and the non-symbiotic Springer’s damselfish (Chrysiptera springeri) which is a close relative. The PCR yielded some products which are bright bands in the gel below (Figure 1). The brighter the bands the more concentration of DNA, showing successful replication. These samples were then cleaned up and sent off for sequencing. The sequencing results were crosschecked with the NCBI database and matches that appeared were not of SIAT7. Instead they matched to other genes like protein FAM20A isoform X3 inform the Southern pig-tailed macaque or monkey (Macaca nemestrina) (Figure 2) or to bacterial genes like protein A2680_02525 from the bacteria, Candidatus kaiserbacteria . These sequencing results are the DNA of one of the bands from the failed attempt using degenerate PCR. These results indicate that our DNA in the degenerate PCR was not successful at producing a partial part of the SIAT7 gene.
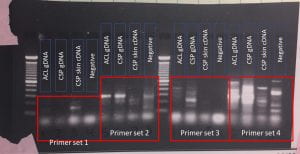
Figure 1. Gel showing the results of degenerate PCR searching for SIAT7 in springers damselfish (chrisyptera springeri, CSP) and clarks clownfish (amphiprion clarkia, ACL) genomic (gDNA) and skin cDNA samples.

Figure 2. Sequencing results and BLASTx alignment for a sample. The BLAST results show a match with the protein FAM20A isoform X3 with the Southern pig-tailed macque (Macaca nemestrina) which is a medium-sized monkey.
Since the degenerate PCR primer was not successful at yielding a partial sequence for SIAT7, another approach to obtaining this sequence was taken. Marian Wahl, a graduate student in Dr. Robert Drew’s lab, had recently sequenced transcriptomes from several species of anemonefish and non-symbiotic fish. Transcriptomes are all of the RNA that is made by genes of an organism. This is of interest because it shows exactly what is made and what will potentially be translated to proteins. This was not available in the spring but became available early this summer. I redesigned primers for four species of anemonefish (list species) and non-symbiotic fish (list species) to be used in the PCR. This provided me with a better chance of getting PCR product because the primers used in the PCR were designed from the exact species they would be testing in. Also, I would be able to see right away if SIAT7 was really present in the fish species because I would be checking their transcriptomes to see if it was present or not. If SIAT7 was present, I would get a gene sequence from the transcriptomes.
To do this a reference gene was identified from the bicolor damselfish (Stegastes partitus; XP_008298796.1). This reference gene was used to find matching sequences from the transcriptomes of the study species using Local BLAST. I found that SIAT7 appeared in all species transcriptomes that were checked. From this, I could say that SIAT7 is found in both symbiotic and non-symbiotic species of fish. However, the specific tissue or tissues it is expressed in and to what extent was not known from this information.

Figure 3. PCR Results using primers designed for Amphirion clarkii (ACL) species and Amblyglyphidodon curacao (AmCu) species.
After going back to look at the specific gene sequences that were used to make the primers, there was evidence that SIAT7 across these species may be paralogs. Paralogs are genes that have evolved by duplication events, resulting in two copies of the gene in different locations of the genome. After duplication, these copies evolve independently, accumulating different mutations. After a long period, these paralogs may still encode for the same protein but can have very different DNA sequences. This is interesting to note because it could be evidence that clownfish symbiosis caused this duplication event to occur. We found paralogs in the Clarks clownfish but not in the other three species we tested which were the staghorn damselfish (Amblyglyphidodon Curacao), the three spot damselfish (Dascyllus Trimaculatus) and the three stripe damselfish (Dascyllus Aruanus). We found this out by aligning the different transcript that were gotten from the Local BLAST. When aligned I found that the Clarks clownfish transcripts with similar trinity numbers (numbers that appear after the letters “DN” in Figure 4) were more closely related than the ones with dissimilar numbers. If the sequence used to make the primers were made using one paralog, other paralogs will not be accounted for in the study and the PCR will not yield consistent results.
To account for paralogs, some bioinformatics was done to identify exactly where duplication events might have occurred and in what species. To do this, transcriptomes for the species of interest were identified and aligned to each other using computer programs such as MUSCLE, TranslatorX, and the NCBI Blast Website (Figure 4). When transcriptomes are aligned, the programs will put similar sequences together and dissimilar sequences further apart from each other. This figure highlights that species with the same sequences (samples with the same Trinity numbers) may be from the same gene. For example, the sequences DN83440 and DN182523 from A. clarkii are probably paralogs but there are two copies of DN182523 which are probably splice variants or have alternative transcription start sites.
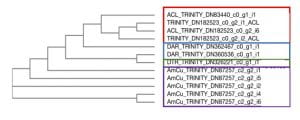 Figure 4. Phylogenetic tree of SIAT7 cDNA samples from Clark’s Clownfish (ACL), Three stripe domino damselfish (DAR), Three spot domino damselfish (DTR) and staghorn damselfish (AmCu).
Figure 4. Phylogenetic tree of SIAT7 cDNA samples from Clark’s Clownfish (ACL), Three stripe domino damselfish (DAR), Three spot domino damselfish (DTR) and staghorn damselfish (AmCu).
There is still much to do so I am continuing work on this project this fall. This figure will be updated to include some well-studied fish and re-rooted to provide more accurate results. Some cichlid fish are more well understood in the evolution of fish, and using these as references for our SIAT7 sequences, can provide me with some information on paralogs. Once paralogs are completely identified, more specific primers can be designed that will hopefully yield consistent PCR results. Another approach that will be taken is to align protein sequences. Right now, the aligning that has been done has used cDNA sequences.
§
From the work done this summer, I can say that SIAT7 is found in symbiotic and non-symbiotic fish that I studied, indicating that clownfish did not lose SIAT7 as part of the evolution of symbiosis with sea anemones. However, I detected evidence of gene duplication which introduced paralogs. Going forward, I seek to understand when these duplication events occurred and if it is related to the clownfish-sea anemone symbiosis. I am looking to seeing if the evolution of paralogs in SIAT7 allowed anemonefish to live symbiotically with anemones or if it is completely unrelated to this.
§
This summer research experience has provided me with an opportunity to continue with a long-term research experience. I stepped out of my comfort zone and experienced new things in the lab and learned immensely from bioinformatics alone. Being able to get results from looking at gene sequences and databases on local computers, and searching national gene databases, I could answer one of my research questions without even picking up a pipette. As a biology student, I underestimated the wealth of information bioinformatics shows and how important it is to do these steps in research. Conducting experiments in the lab is rewarding but interpreting the data, and understanding it is the main goal. This summer research experience, I learned to think about long term goals and the bigger picture. Having participated in only short-term research experiences before, I was usually just thrown into a situation where I had to think quickly on my feet and do a series of experiments and interpret my immediate results. However, being at UMass during the summer, I could continue work I had started before. This allowed me to see what a long-term project entails. Data interpretation and relating results to a goal is something that I have strongly developed this summer. I feel much more prepared to pursue more long-term projects. I have developed myself as a critical thinker and a troubleshooter in my research and found a new appreciation for the study of bioinformatics.