Progress Report on the Creation of a Microfluidic Device for the detection and Characterization of Exosomes
By Ken-Lee Sterling
Collaborators: Michael Nessralla, Vinh Phan, Jenny E Luo Yau and Prof. Milana Vasudev

Portrait of Ken-Lee Sterling at work in his lab.
Introduction
During the last few decades fatal illnesses such as cancer seem to have become more prevalent in undeveloped as well as developed societies. The tools at our disposal to fight these diseases have become increasingly vast (endnote 1). However, detection and prevention are much more beneficial and productive than attempting to combat the cancerous cells. This leads to the question of if a pre-cancerous formation could be detected before it reaches the point of needed invasive combat stage.
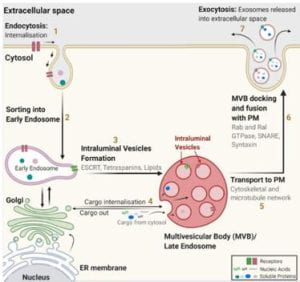
Figure 1. Exosome structure and origin
Through the development and creation of a Microfluidic device with an implanted SERS detector it could be possible to present the exosomes to the embedded sensor for real-time characterization and detection. This could theoretically be a less expensive and affordable method of cancer detection and prevention. If the microfluidic device is easy to make and repeatable to a high degree, that will allow testing with more accuracy and less variation. When an appropriate outlet design and channel shape are incorporated, then the particles can be captured with high purity, high yield, and at a high rate concerning the concentration of the solution. This allows for downstream analysis, which in this project correlates to a SERS sensor (endnotes 2,3) which will be used to analyze the particle.
A microfluidic chip is a pattern of molded or engraved microchannels/ pathways. The network of microchannels can be connected and incorporated into a macro environment4. Microfluidic devices use the unique physical and chemical properties of liquids and gasses at the micro and nano scale. The most studied way to control the fluids is the use of custom shaped and directed micro channels5. Channel shapes can focus, concentrate, order, separate, transfer, and mix the particles and fluids (endnotes 5,6).
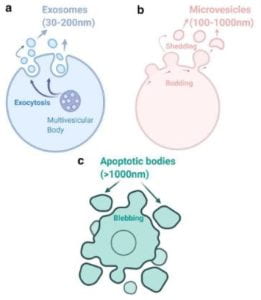
Figure 2. Descriptive images of different types of cellular vehicles
Exosomes, otherwise known as extracellular vehicles (EVs) are classified into three groups based on their size and biogenesis (footnote 7). Exosomes range from (30-200nm) to micro vesicles (100-1000) and apoptotic bodies (>1000nm) (Fig 2) (endnotes 8, 9). Exosomes are of endocytic origin (3,1), which means that they arise from the intake of material into the cell through the folding and subsequent encapsulation of the lipid membrane around the materials (Fig 1).7 EVs can be further categorized based on their density, composition, and function. EVs are membrane-bound due to their nature of being carriers of cell-cell communication. They take on a spherical shape and consist of proteins such as CD9, CD63, and CD81, which are part of the Tetraspanin family and cytoskeletal components. These vesicles, once secreted can provide key information from the cell of origin, like a “cell biopsy.”
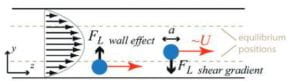
Figure 3. Effect of Channel Shape and Size on particle movement
To understand the device, the physics that drive the device must be understood. Microfluidic devices use the unique chemical and physical properties of liquids and gasses at the micro and nano scale (endnote 5). The most studied way to control the fluids is using shaped channels. Channel shapes can focus, concentrate, order, separate, transfer, and mix the particles and fluids (endnote 6). A deviation from a straight channel introduces dominant/weaker lift forces and internal lift through the interaction with the particle and the adjacent wall (Fig 3). Focusing the particles and fluid into specific shapes and channels allows the particles to self-sort and filter. Using a square channel as an example, randomly dispersed particles of a certain size will focus on four symmetric equilibrium positions near the center of the channel wall face (Fig 4 a). When an appropriate outlet design and channel shape are incorporated, then the particles can be captured with high purity, high yield, and at a high rate concerning the concentration of the solution. This allows for downstream analysis, which in this project correlates to a SERS sensor (endnote 10). The SERS sensor will be used to analyze the particles.
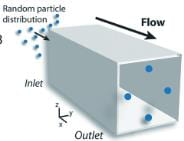
Figure 4.a. Particle orientation within a square tube on indeterminate length
The ultimate objective of the entire apparatus is to seamlessly integrate a Surface Enhanced Raman Sensor (SERS) into the microfluidic device and enable the fluid to flow through the sensor, thus facilitating the identification of exosomes. A reservoir will hold a solution of PBS buffer and 1% Bovine, in which the exosomes will be suspended. Using a connected pump, the fluid will flow from the reservoir through the microfluidic device and get filtered before passing in front of the SERS sensor, which will help detect the exosomes. This will allow the detection of Ovarian Cancer exosomes, which can confirm or deny a diagnosis of ovarian cancer in a woman. Early diagnosis is essential for finding cancer cells. The traditional and current methods (diagnostic magnetic resonance imaging (MRI) and computed tomography (CT) are typically highly costly and come with several downsides. A high dosage of radiation in the long term can cause damage to healthy cells and may cause serious issues for the patient depending on the cells that are affected or not. The device is a rapid, non-invasive method that will allow for rapid cancer diagnosis. Notably, the device will have characteristics that improve on similar devices in the category, which are discussed in length below.
Methods/ Technical Approaches
During the initial discussions of the PDMS casting, the consensus was that different PDMS to curing agent ratios had to be synthesized to determine which ratio would yield the best overall results. Calculations were done to determine the proper breakdown of the PDMS to curing agent ratio. The initial casting dimensions were based on version 15 of the solid works models (Fig.4.b). The proper breakdown of the ratios was calculated through the simple equation of .Where the being the total internal volume of version 15 of Solid Works model. is the total number of parts. The total internal volume of version 15 of the PDMS mold was calculated to be 3.4 mL.
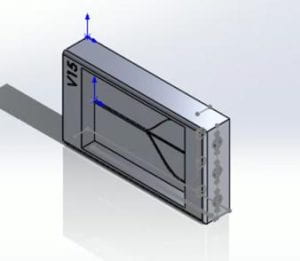
Figure 4.b. Version 15 of the microfluidic device
With the z-height being 0.5 cm, the x-height being 3.4 cm, and the y-height being 2.00 cm, resulting in a total volume of 3.4 cm3 or 3.4 mL. It was decided that the ratios that would be tested and cast would be 10:1 and 15:1. The total amounts of the required volumes for both the 10:1 and 15:1 were calculated using the equation previously mentioned. For the 10:1 and 15:1 casting, there was an assumed 0.1 mL margin of error for the castings and potential residue material that would be left behind from mixing the PDMS/Curing agent to the transfer into the models. The calculations for the 15:1 casting proceeded with a total of 16 parts being assumed, with 15 parts being PDMS and 1 part being the curing agent 3.5 mL )16=0.2187 mL 0.2187 mL∙15=3.2812 mL PDMS, 0.2187 mL curing agent. For the 10:1, the calculations were done similarly where ten parts were assumed to be the PDMS and 1 part was assumed to be the curing agent for a total of 11 parts resulting in the final equation being 3.5 mL 11=0.318 mL, 0.318∙10=3.181 mL PDMS, 0.318 mL curing.


Fig. 5. The results of the casting with the 15:1 and 10:1. The initial models were cured for roughly 48 hours. Even after the 48 hours recommend curing time the PDMS molds were still incredibly unstable.
After the initial casts of the 15:1 and 10:1 mold, it was realized that the ratios of PDMS in the mixture resulted in very unstable and structurally weak molds (Fig.5). At this point in the experimental process the molds were still curing in standard room temperature, anywhere from 20-23 degrees Celsius. After it was determined that the current ratios of PDMS-to-curing agent ratios that were currently being used resulted in inadequate and unstable molds, the conclusion was made that the next sample of molds would be done in accordance with the following ratios, 11:1, 11:2, 10:1, and 10:2. Between recasting new PDMS molds the B9 printer was in need of a recalibration. Since the project was in the later stages of physical development, the decision was made that the B9 printer should be recalibrated to the desired resolution of 50 μm. The 11:1, 11:2, 10:1, and 10:2 molds were removed and examined. When the molds were released from the casts the 10:2 PDMS casts were noticeably softer and more malleable than the 11:2 casts (Fig. 6B). There also appeared to be signs of PDMS residue left behind upon removing the PDMS casting mold (Fig. 6A, D).




Fig 6 (A.B.C.D: top to bottom, clockwise): The results of the different PDMS curing ratios after the PDMS had been removed.
Upon the realization that the PDMS was stuck to the foundation of its mold during removal, the team made the decision to use mold release and a control group of no mold release on the casts themselves. The team made the decision that based on our previous casts we would utilize the 11:2 ratio PDMS mixture-it was the most structurally sound. On February 11th, 2023, the PDMS a new set of molds were produced 3 casts were done using mold release and 3 were done using the coconut oil. Due to the fact the oven could not be used to increase the curing time these samples were left to cure for 120 hours. Even after the 5-day curing time the molds did appear to be structurally weak (Fig.5). The way in which we have approached the current methods in casting and producing this device align with the current goals of keeping this device reusable and inexpensive.

Fig. 7. Isometric view of V6 Device

Fig. 8. Isometric View of version 11 of the device.
We have been able to create numerous models using the PDMS with little cost. We have also incorporated the technique of washing the PDMS casting trays using the chemical compound known as hexane(s) C6H14. Since hexane was utilized as a washing method-to dissolve the PDMS from the trays has allowed for the reuse of many of the casting trays and keep the costs of printing down. The cost of materials and financial use has been kept to a minimum during the project to a minimum by using low amounts of the PDMS cast silicon base and the caring agent. Based on the calculations previously mentioned, we do not use more than four grams at a time of the PDMS casting and curing agent combined. With a total of six trays for potential casts, there are no more than 24 grams used out of the 200-gram base and 20-gram curing combined.
Device Design Updates
Our previous casts we would utilize the 11:2 ratio PDMS mixture-it was the most structurally sound. On February 11th, 2023, the PDMS a new set of molds were produced 3 casts were done using mold release and 3 were done using the coconut oil. Due to the fact the oven could not be used to increase the curing time these samples were left to cure for 120 hours. Even after the 5-day curing time the molds did appear to be structurally weak (Fig.5). The way in which we have approached the current methods in casting and producing this device align with the current goals of keeping this device reusable and inexpensive.

Fig. 9. Top view of version 12 of the device
We have been able to create numerous models using the PDMS with little cost. We have also incorporated the technique of washing the PDMS casting trays using the chemical compound known as hexane(s) C6H14. Since hexane was utilized as a washing method-to dissolve the PDMS from the trays has allowed for the reuse of many of the casting trays and keep the costs of printing down. The cost of materials and financial use has been kept to a minimum during the project to a minimum by using low amounts of the PDMS cast silicon base and the caring agent. Based on the calculations previously mentioned, we do not use more than four grams at a time of the PDMS casting and curing agent combined. With a total of six trays for potential casts, there are no more than 24 grams used out of the 200-gram base and 20-gram curing combined.

Fig 10. PDMS castings conducted on 2/16/23 where no mold release was utilized.
This section provides a detailed analysis of the advancements in device design and highlights the potential benefits and drawbacks of each advancement. Starting with Version 6 (Fig.7) the device channels and the fluid manifold were the main development. Looking at Figure 7 you can see through the translucent top piece the 3 channels sized to focus 30, 50, and 75nm exosomes. The research focus has shifted towards exploring different ratios of PDMS, in conjunction with varying mold releases and ratios. To enable this, a series of molds were developed that allowed for testing of various combinations of base-to-curing ratios, temperature, and time in the oven. To reduce material usage and accommodate size constraints, the mold size was minimized, and the channels were simplified to only 50 nm.

Fig. 11. Another PDMS casting that was done on 2/16/23/ Left side, was with no use of any type of pf mold release. Right side, with the use of mold release
When deciding between a negative mold (or reverse mold), which produces a negative impression of an object or pattern, and a positive mold (or direct mold), which produces a positive impression, we opted for the latter to create the microfluidic device. The process of creating a positive mold involves multiple steps, starting with mixing the base and curing agent in a specific ratio in a separate dish. The material is then poured into the mold, and air bubbles are removed either through vacuum or manually. Lastly, the mold is placed in the oven for a specific amount of time at a specific temperature.
After the mold material has hardened, it is removed from the object or pattern, revealing a positive impression of the original. The casting material fills the positive space of the mold, taking on the shape of the original object or pattern, resulting in a replica or a copy of the original. Positive molds are an efficient and cost-effective solution for creating multiple copies of an object or pattern for a wide range of applications. Versions 11 and 12 (Fig.8,9) continued the trend of incremental improvements in the mold design, while simultaneously reducing the weight of the mold itself, thus decreasing material costs. Design simplifications enabled the team to increase the effective casting area, further optimizing the device. However, during testing with thinner molds, air bubbles were observed forming on the bottom of the cast. This issue was attributed to an uneven heat distribution across the different regions of the mold. To address this problem, the team decided to keep the 5 sides of the casting mold at a uniform thickness moving forward. As previously mentioned, the ideal ratio of PDMS to curing agent was found to be 11:2, followed by a curing time of 4 hours at 50 °C, which produced the best results. Version 13 onwards, the focus shifted towards developing a functional device for testing and data analysis purposes. To achieve this objective, the team procured the GENIE Touch Syringe Pump platform from PI for precise fluid manipulation and received specialized training on the HIROX lab microscope for obtaining high-resolution images of the device during operation. While the device design is being fine-tuned and made watertight, initial observations are being carried out under a standard lab bench microscope.
Results
So far in the experimental process several microfluidic channel prototypes have been synthesized. Due to variables that have not yet been identified it has been difficult to determine what the causes of the differences of the results were. Figure 10 is an example of a cast that was conducted on February 16,2023. This PDMS was casted with no use of any type of casting mold release. Whereas in figure 11, the right-handed cast was done with the use of mold release. Through these two different samples, we concluded that the mold release in combination with the PDMS had this interaction that prevented the PDMS from fully curing. This effect is more noticeable in figure 12. On February 12, 2023, casts were also conducted. However, these results were profoundly different form the casts that were later done on the 16th. After the initial casts using the mold release, another casting was done to confirm the idea that mold release effected the structure of the PDMS (fig.11). We have however been able to determine that through our casting technique we have been able to maintain some level of resolution. Through a microscope the resolution require has been somewhat confirmed (fig.13). Even though the casting techniques have yet been perfected. The concept is there, and we have been able to produce micro channels.

Fig. 12. Two Casts that were conducted with the use of mold release.
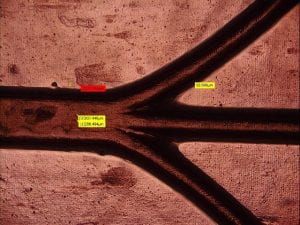
Fig. 13. HiRox microscope image of the microfluidic channels.
Endnotes
1 Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(1):7-34. doi:10.3322/caac.21551
2 Perumal J, Wang Y, Attia ABE, Dinish US, Olivo M. Towards a point-of-care SERS sensor for biomedical and agri-food analysis applications: a review of recent advancements. Nanoscale. 2021;13(2):553-580. doi:10.1039/d0nr06832b
3 Lee C, Carney R, Lam K, Chan JW. SERS analysis of selectively captured exosomes using an integrin-specific peptide ligand. Journal of Raman Spectroscopy. 2017;48(12):1771-1776. doi:10.1002/jrs.5234
4 Team E. Microfluidics: A general overview of microfluidics. Elveflow. Published online February 5, 2021. Accessed September 14, 2022. https://www.elveflow.com/microfluidic-reviews/general-microfluidics/a-general-overview-of-microfluidics/
5 Kim U, Oh B, Ahn J, Lee S, Cho Y. Inertia–Acoustophoresis Hybrid Microfluidic Device for Rapid and Efficient Cell Separation. Sensors. 2022;22(13):4709. doi:10.3390/s22134709
6 Amini H, Lee W, Carlo DD. Inertial microfluidic physics. Lab Chip. 2014;14(15):2739-2761. doi:10.1039/C4LC00128A
7 Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. 2021;19:47. doi:10.1186/s12964-021-00730-1
8 Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514. doi:10.1146/annurev-biochem-013118-111902
9 Lee C, Carney R, Lam K, Chan JW. SERS analysis of selectively captured exosomes using an integrin-specific peptide ligand. Journal of Raman Spectroscopy. 2017;48(12):1771-1776. doi:10.1002/jrs.5234
10 Perumal J, Wang Y, Attia ABE, Dinish US, Olivo M. Towards a point-of-care SERS sensor for biomedical and agri-food analysis applications: a review of recent advancements. Nanoscale. 2021;13(2):553-580. doi:10.1039/d0nr06832b